|
||
|
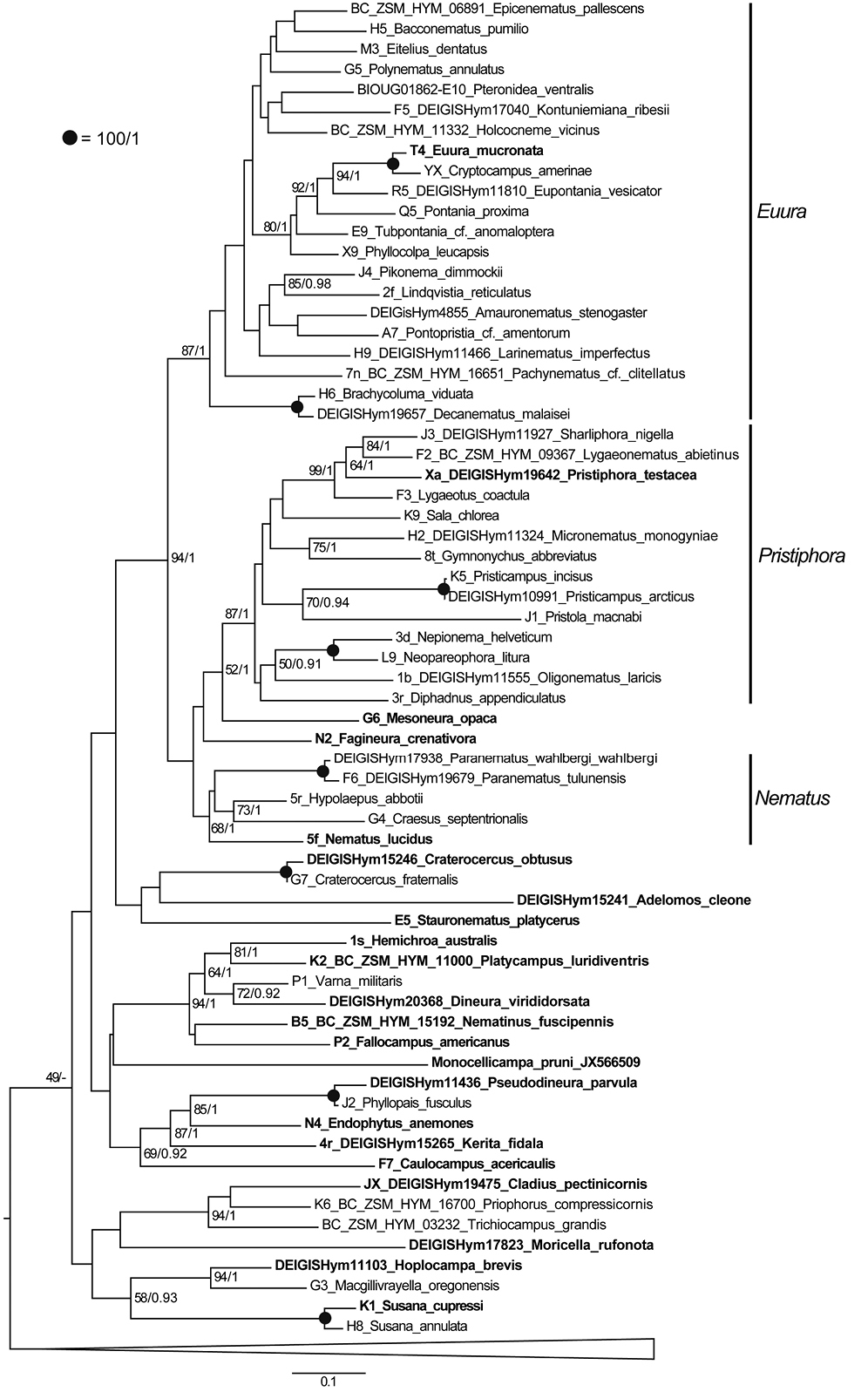
Phylogenetic relationships of type species for which at least 400 bp of the CoI barcode region was available. The tree was reconstructed according to a ML analysis allowing a separate GTR+I+G4 model of substitution for each gene. Numbers above or right of branches show bootstrap proportions (%, BP) followed by posterior probabilities (PP) from the corresponding Bayesian analysis. Branches receiving maximum support (BP=100%, PP=1) are denoted by a black dot. Support values for weakly supported branches (PP<0.9 and BP<50) are not shown. Nematinae was not monophyletic in the Bayesian analysis, because Moricella rufonota (only 658 bp of CoI available) was weakly placed as sister to the Strongylogaster+Nesoselandria clade. Some terminals in the dataset are composites of two conspecific specimens (indicated by two ID numbers, for example J3 and DEIGISHym11927 or F2 and BC ZSM HYM 09367). Four non-type species (Pristicampus incisus, Paranematus tulunensis, Craterocercus fraternalis, and Susana annulata) were also included in the analysis, because the CoI or NaK (for S. cupressi) sequences available for the type species of the respective genera were not sufficient to place them reliably in the tree. Type species in bold are the basis for genera defined here. Outgroup taxa have been collapsed for simplicity. Voucher ID numbers (e.g. F2) correspond to specimen identifiers in previous phylogenetic trees with different species names. The scale bar shows the number of estimated substitutions per nucleotide position. |