|
||
|
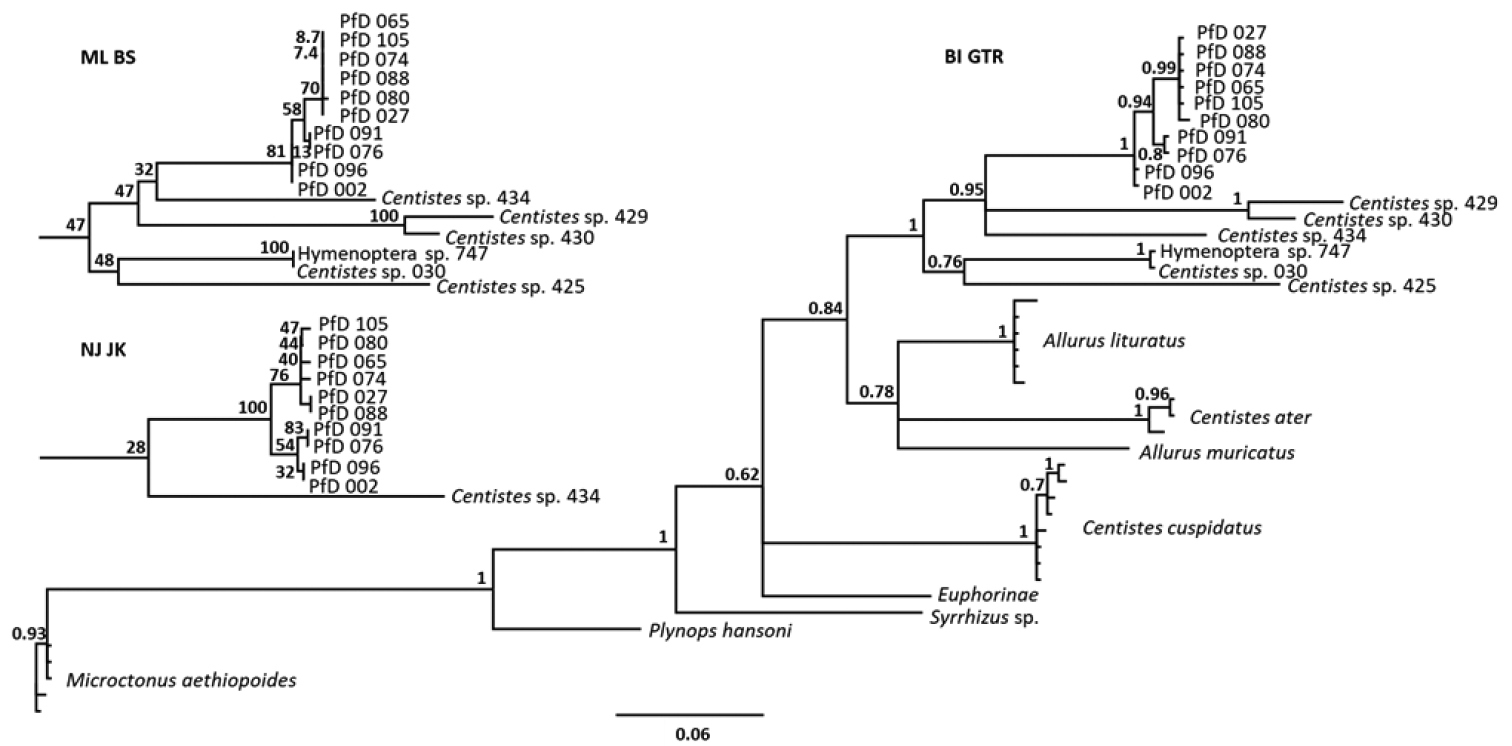
Phylogenetic analyses using CO1 sequences from parasitoids in D. undecimpunctata aligned with the most similar sequences from NCBI. Complete tree is BI using the GTR nucleotide substitution model with PP branch support, subtrees show results from the NJ (JK support) and ML (BS support) methods to highlight the similarities in resolution between methods in relation to the samples generated in this study. All analyses used the same MAFFT alignment (Suppl. material 1). Complete trees for all methods are included in Suppl. material 1: Figs S1–S5. Numbers after species names refer to the last three digits of the GenBank accessions, all other numbers refer to internal lab accession numbers which can be cross referenced in Table 1. |