(C) 2013 Adrien Perrard. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
The only previous comprehensive phylogenetic analysis of the 22 species of the genus Vespa was based on just 11 morphological characters and resulted in only limited resolution. In order to improve the phylogenetic inference, we carried out a simultaneous analysis with 45 morphological characters and data from four mitochondrial and two nuclear genes. The results support a number of the previously found relationships. The monophyly of the genus Vespa and the existence of a main clade excluding Vespa basalis and Vespa binghami are confirmed. The tropica group is supported. The affinis group is not supported; molecular data relate the previously unresolved Vespa orientalis to Vespa affinis + Vespa mocsaryana.
Hornet, Phylogeny, Total Evidence, Vespidae
The genus Vespa is one of the four genera of the subfamily Vespinae; it is composed of 22 extant species of hornets (
The first cladistic study of Vespa was that of
Archer’s study was based mostly on male characters, which are known to be reliable phylogenetic characters (e. g.
This study presents results from an ongoing project on the evolution of vespine wasps, focusing on the genus Vespa. Our aims are to confirm the monophyly of the genus Vespa, a question not addressed by
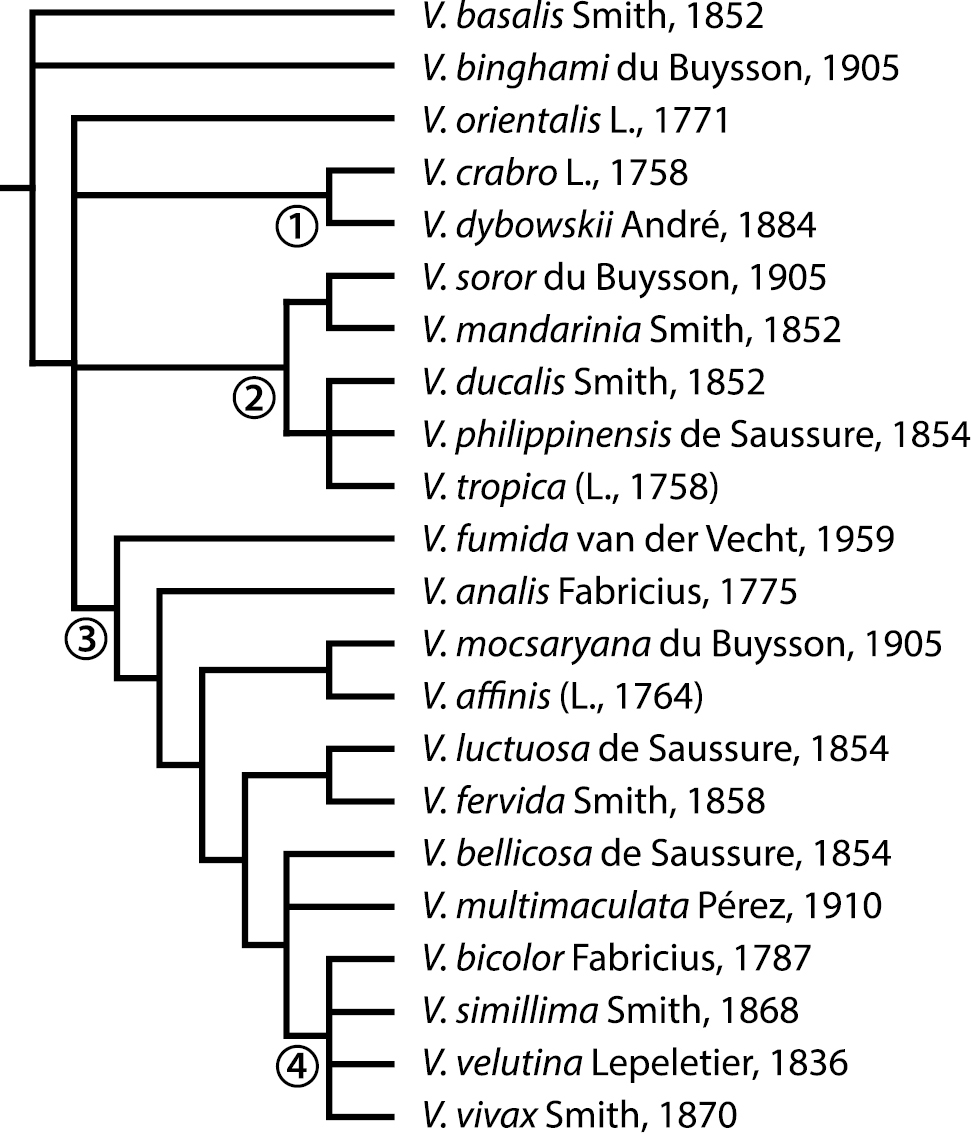
Phylogeny of the genus Vespa after
Phylogenetic relationships were inferred for the 22 species of the genus Vespa and five other species of Vespidae as outgroup: Dolichovespula media (Retzius, 1783), Provespa anomala (de Saussure, 1854), Provespa barthelemyi (du Buysson, 1905) and Vespula germanica (Fabricius, 1793) from the subfamily Vespinae and Polistes dominula (Christ, 1791) from the Polistinae.
The morphological matrix was scored from specimens in the following natural history collections: American Museum of Natural History, Muséum National d’Histoire Naturelle, Nationaal Natuurhistorisch Museum and United States National Museum of Natural History. Specimens for molecular study were collected by J.K. and J.M.C. and preserved in 95 - 99% ethanol.
DNA was extracted from one leg and one antenna per specimen using QIAGEN “DNeasy tissue Kit”. Genes were amplified using PCR with PuReTaq Ready-To-Go beads in a total volume of 25µL including primers (Appendix 1) and DNA. Amplification cycles were specific to genes (Appendix 2). AMPures and CleanSEQ procedures were used for DNA purification and sequencing was performed on ABI PRISM 3730xl machines by Agencourt Biosciences (Beverly, USA). One missing gene fragment of Vespa basalis was obtained from Genbank database (accession number: AB585949).
The analyses are based on 45 morphological characters and multiple nuclear and mitochondrial loci, comprising: 374 sites of 12S, 528 sites of 16S, 2231 sites of 28S (sequenced in 4 fragments), 1442 sites of CO1 (sequenced in 2 fragments), 880 sites of elongation factor 1α (EF1α), and 328 sites of H3. Each of these genes was aligned separately using the MAFFT software with the L-INS-i algorithm (
Phylogenetic analyses were performed in a parsimony framework using TNT (
Analysis of the 45 morphological characters (Appendix 3) resulted in a single most parsimonious tree (not shown; Length: 107, Consistency Index (CI): 0.617, Retention Index (RI): 0.784). The support tree is shown in Fig. 2. The only difference is that the most parsimonious cladogram resolves Vespa orientalis as the sister species to the clade composed of mandarinia + soror and the affinis group; the support tree (Fig. 2) does not include this node. On both trees the genus Vespa appears as monophyletic, and Vespa basalis the sister species of the rest of the genus, followed by Vespa binghami. The 20 remaining species are grouped in three clades, relationships among which are not resolved (Fig. 2): a clade with only Vespa orientalis, the second comprising crabro + dybowskii together with Vespa tropica and its two closely related species, and the third clade with the remaining 14 species. The tropica group sensu
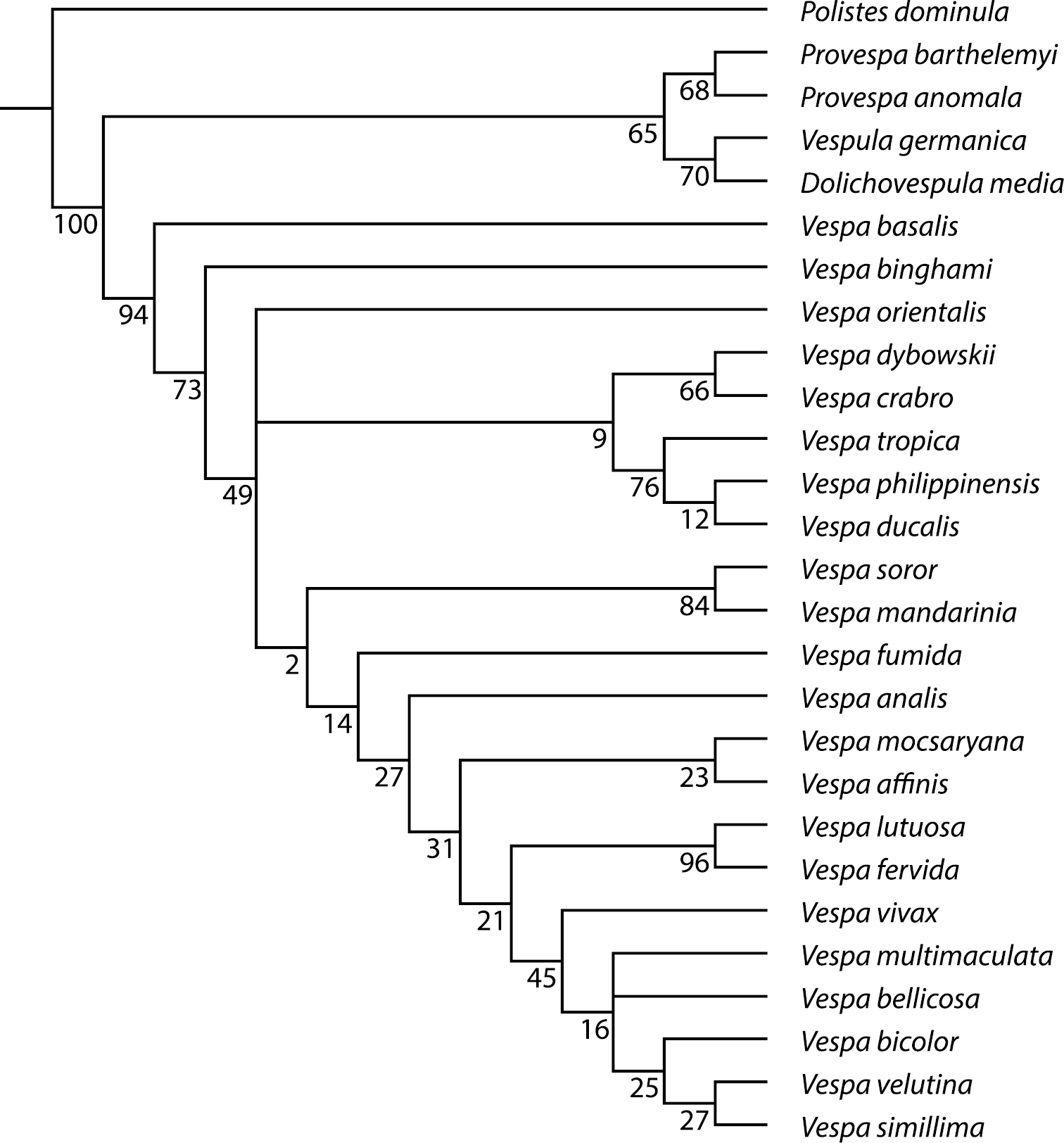
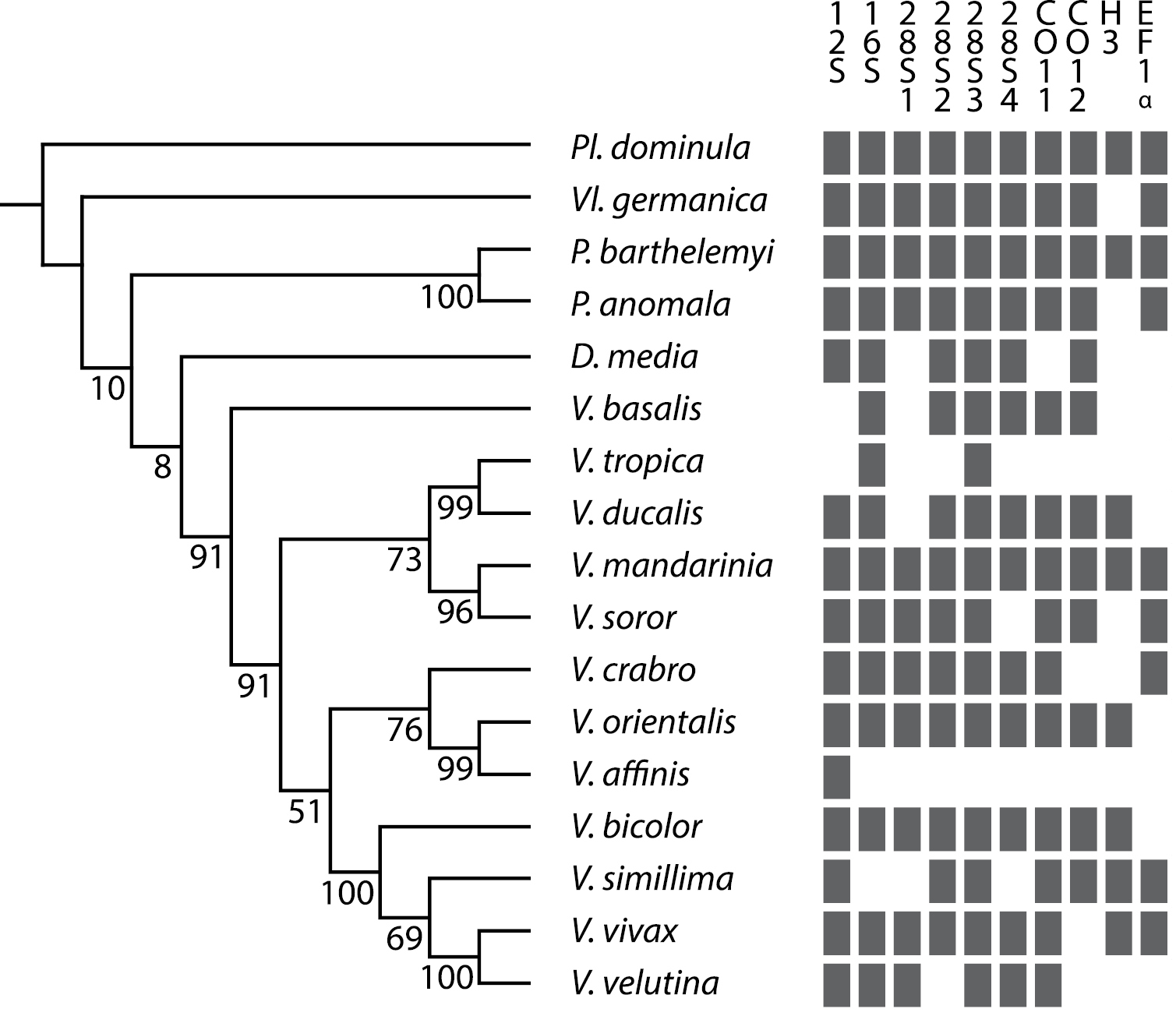
Of the 27 studied species, specimens relatively recently collected were available for 17 species (including the outgroups). Due to low quality DNA templates and the use of non-specific primers, molecular data are not homogeneous across the genus. The monophyly of the genus Vespa was found in most analyses based on single genes and in the analysis of merged alignments (Table 1, Fig. 3). The main species-groups appear monophyletic over most analyses except for the low variation 28S. Vespa basalis remains the basal species in the genus. Two main clades diverge in the remaining species:the monophyletic tropica group is the first clade, while the bicolor group and a clade of Vespa crabro, Vespa mocsaryana and Vespa affinis form the second. The bicolor group sensu
Support tree for relationships among the 22 Vespa species based on 45 morphological characters.Supports for nodes are given in GC-values (see text for explanation) when they are greater than zero.
Results of phylogenetic analyses of molecular data for the genus Vespa. Marker: gene used in the analysis. “Multi-locus” marker is a combination of all genes used. N tree: number of most parsimonious trees. Length: length of the most parsimonious trees. Vespa, bicolor, mandarinia, tropica: monophyly of the considered clade when it is tested.
| marker | N tree | length | CI | RI | Vespa | bicolor | mandarinia | tropica |
| 12S | 5 | 261 | 0.709 | 0.651 | yes | yes | yes | - |
| 16S | 2 | 332 | 0.654 | 0.545 | yes | yes | yes | yes |
| 28S | >1000 | 145 | 0.909 | 0.750 | no | yes | no | no |
| CO1 | 1 | 1431 | 0.508 | 0.369 | yes | yes | yes | - |
| EF1a | 1 | 219 | 0.886 | 0.819 | yes | yes | yes | - |
| H3 | 1 | 81 | 0.889 | 0.690 | no | yes | - | - |
| Multi-locus | 207 | 2494 | 0.620 | 0.468 | yes | yes | yes | yes |
Support tree for the relationships among 13 Vespa species based on the six genes. Supports for nodes are given in GC-values. Grey rectangles show the molecular markers available for each species.
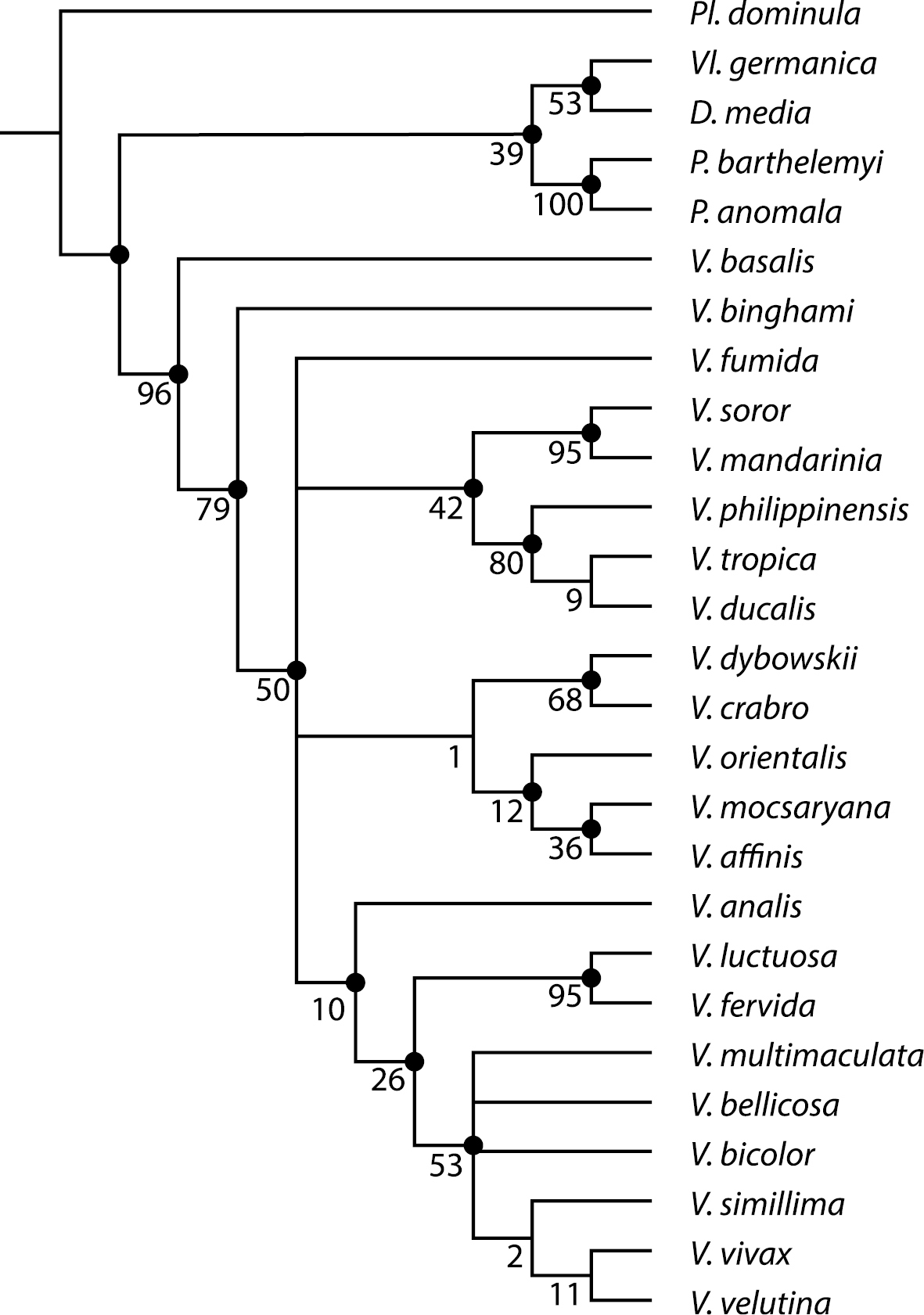
The combined analysis of morphological and molecular data returned eight equally parsimonious trees all showing Vespa monophyletic with Vespa basalis as the sister species of the rest of the genus (not shown; Length: 2665, CI: 0.608, RI: 0.488). The consensus tree (not shown) is completely resolved except for a node including the bicolor group, Vespa bellicosa and Vespa multimaculata. In the support tree (Fig. 4), two internal nodes are collapsed, because of a different position of Vespa fumida.
The monophyly of the genus is supported by five synapomorphies: the long prestigma, the developed vertex, the strongly elevated interantennal space, the presence of a carina on the hindcoxa, and the projection at the apex of the digitus in males.
The addition of molecular data to the morphological matrix resulted in stronger support of the tropica group and resolved the position of Vespa orientalis, as part of a clade with two morphologically different species, Vespa affinis and Vespa mocsaryana, close to Vespa crabro. The affinis group sensu
In the combined analysis tree, most of the clades of Vespa species are supported by morphological characters, five of which are uncontroverted synapomorphies. Vespa basalis is distinguished from the other species on the basis of the aedeagal apical lobes in males and the edges of the interantennal space. The main clade excluding Vespa basalis and Vespa binghami is supported by the clypeal punctures dense mesally in females and the emarginated apical margins of the metasomal sterna VI and VII in males.
Within the main Vespa clade, the tropica group is morphologically supported by the triangular apico-lateral angles of female clypeus only, but molecular data confirms this homology. Within this group, Vespa mandarinia + Vespa soror is defined by two uncontroverted characters: the spade-shape of the aedeagus apex in males and the expansion of the gena behind the eyes. The three other species of the tropica group share the presence of pronotal striae, long first metasomal segment and marked scutal and metapleural punctures.
The two species of the crabro group share two secondary reversions: the straight apical margin of the male metasomal sternum VI and the loss of digital apical process. Vespa affinis and Vespa mocsaryana share the posteromedially deeply emarginated male metasomal sterna VI and VII and long first metasomal segment. The clade consisting of Vespa orientalis and Vespa affinis + Vespa mocsaryana is supported by the short malar space, a homoplastic synapomorphy found also in the clade of Vespa analis, Vespa luctuosa + Vespa fervida, Vespa multimaculata, Vespa bellicosa and the bicolor group. This latter clade is also supported by the bulbous aedeagal shaft in males. The clade of these species but Vespa analis is morphologically supported by a pretegular carina ventrally effaced, the presence of a few ventral striae on the female pronotum and the apical margin of the male metasomal sternum VII deeply emarginate. Vespa fervida + Vespa luctuosa is supported by the well defined punctures on metapleura and lateral faces of the metasomal tergum II as well as the uncontroverted synapomorphy of the median process in the apical margin of the metasomal sternum VII. Finally, the clade of the bicolor group and Vespa multimaculata and Vespa bellicosa is supported by the distinct interruption of the pronotal carina by the pronotal pit. The four remaining clades within the genus Vespa are not diagnosed by morphological characters. These latter clades also have low support under symmetric resampling (Fig. 4).
Support tree for the relationships within the genus Vespa based on a combined analysis. Support tree based on 45 morphological characters and six genes. Black nodes indicate clades supported by morphological characters. Absence of mark on nodes indicates clades diagnosed by molecular data only. Supports for nodes are given in GC-values.
Our analyses of both morphological and molecular characters confirm the monophyly of the genus Vespa. This genus was first diagnosed from other Vespidae on the basis of the shape of the head (
While our molecular sample is incomplete, it nonetheless confirms the monophyly of two of Archer’s species groups within Vespa based on the morphology (crabro and tropica groups). Molecular data help to place Vespa orientalis, which both Archer’s and our morphological analyses failed to resolve well. Vespa orientalis is the only Vespa species distributed in arid areas in central Asia and the Middle-East. A close relationship of this species to Vespa affinis + Vespa mocsaryana despite obvious morphological differences begs the question of morphological adaptations to arid climates in Vespa orientalis that may have blurred the morphological phylogenetic signal. However, the close relationship of Vespa orientalis and Vespa affinis is suggested only by the 12S gene in the molecular data. Further gene sequences for this last species and for Vespa mocsaryana are necessary to clarify whether the clade consisting of Vespa orientalis and Vespa affinis + Vespa mocsaryana is definitively supported.
Our results are also consistent with previous authors regarding the close relationships of Vespa luctuosa and Vespa fervida, which are very similar in their morphology (
Archer’s finding of a main clade of Vespa excluding Vespa basalis and Vespa binghami has been confirmed both by morphological and molecular data. Our results also suggest that the nocturnal species Vespa binghami is closer to the main clade of Vespa than is Vespa basalis. Morphological adaptations to nocturnal habits in Vespa binghami such as enlarged ocelli are thus autapomorphies. Recognition of the subgenus Nyctovespa, with Vespa binghami as sole included species (
Our extended morphological matrix and the molecular sequences partly support Archer’s results, and this analysis confirms that male characters such as the shape of the last metasomal sterna and the genitalia are reliable phylogenetic characters in Vespidae.
We thank B. Baliff, and M. Cunningham from the University of Vermont and F. Jacquet from the Muséum National d’Histoire Naturelle for assistance with obtaining the molecular data. This work benefited from NSF grant DEB-0843505. The first author benefited from an Annette Kade Fellowship from the American Museum of Natural History.
Primers used for sequencing the six genes.
| Marker | Primer | Name | Sequence (5’-3’) |
| 12S | F | 12S AI | AAACTAGGATTAGATACCCTATTAT |
| 12S | R | 12S BI | AAGAGCGACGGGCGATGTGT |
| 16S | F | 16S A | CGCCTGTTTATCAAAAACAT |
| 16S | R | 16S B | CTCCGGTTTGAACTCAGATCA |
| 28S-1 | F | 28S 1A | CCCSCGTAAYTTAGGCATAT |
| 28S-1 | R | 28S 4 BR | CCTTGGTCCGTGTTTCAAGAC |
| 28S-2 | F | 28S 3.2a | AGTACGTGAAACCGTTCASGGGT |
| 28S-2 | R | 28S Br | TCGGAAGGAACCAGCTACTA |
| 28S-3 | F | 28S 4a | GGAGTCTAGCATGTGYGCAAGTC |
| 28S-3 | R | 28S 5b | CCACAGCGCCGATTCTGCTTACC |
| 28S-4 | F | 28S 4.8a | ACCTATTCTCAAACTTTAAATGG |
| 28S-4 | R | 28S 7b1 | GACTTCCCTTACCTACAT |
| CO1-1 | F | LCO | GGTCAACAAATCATAAAGATATTGG |
| CO1-1 | R | HCO out out | GTAAATATATGRTGDGCTC |
| CO1-2 | F | Jerry | CAACATTTATTTTGATTTTTTGG |
| CO1-2 | R | Pat | TCCAATGCACTAATCTGCCATATTA |
| EF1α | F | HaF2For | GGGYAAAGGWTCCTTCAARTATGC |
| EF1α | R | F2Rev1 | AATCAGCAGCACCTTTAGGTGG |
| H3 | F | H3-AF | ATGGCTCGTACCAAGCAGACVGC |
| H3 | R | H3-AR | GTCACYATYATGCCYAAGGATAT |
PCR program of each marker with temperature and time (°C – minute). Den. = Denaturing phase. Anneal. = Annealing phase. Elong. = Elongation phase. N = number of cycles of Denaturing + Annealing + Elongation phases.
| Marker | Den. | Anneal. | Elong. | N |
|---|---|---|---|---|
| 12S | 97 – 0.5 | 42 – 0.75 | 68 – 0.5 | 40 |
| 16S | 94 – 0.5 | 42 – 0.5 | 72 – 0.5 | 40 |
| 28S-1 | 94 – 1 | 43.5 – 0.5 | 72 – 1 | 40 |
| 28S-2 | 94 – 1 | 43.5 – 0.5 | 72 – 1 | 40 |
| 28S-3 | 94 – 1 | 43.5 – 0.5 | 72 – 1 | 40 |
| 28S-4 | 94 – 1 | 40 – 0.5 | 72 – 1 | 40 |
| CO1-1 | 94 – 0.5 | 36 / 51 – 0.5 | 72 – 0.5 | 5 / 35 |
| CO1-2 | 94 – 0.5 | 36 / 48 – 1 | 72 – 1 | 5 / 35 |
| EF1α | 94 – 1 | 54 – 1 | 72 – 1.5 | 35 |
| H3 | 94 – 0.4 | 51 – 0.5 | 72 – 0.75 | 40 |
Matrix of morphological characters. Outgroup species are in grey.
| Character | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 4 | 4 | 4 |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 | 1 | 2 | 3 | 4 | 5 | |
| Polistes dominulа | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Dolichovespula media | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 5 | 0 | 2 | 0 | 0 | 0 |
| Vespa germanica | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 3 | - | - | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 6 | 0 | 1 | 0 | 0 | 0 |
| Provespa anomala | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | - | 0 | 2 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 7 | 1 | 1 | 0 | 0 | 0 |
| Provespa barthelemyi | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | - | 0 | 2 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 4 | 0 | 1 | 0 | 0 | 0 |
| Vespa affinis | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| Vespa analis | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 1 | 1 | 0/1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| Vespa basalis | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 2 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 0 |
| Vespa bellicosa | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| Vespa bicolor | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| Vespa binghami | 2 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| Vespa crabro | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 0 | 1 | 1 | 0/1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| Vespa ducalis | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| Vespa dybowskii | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| Vespa fervida | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| Vespa fumida | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 0 | 1 | 1 |
| Vespa luctuosa | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| Vespa mandarinia | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 0 | 3 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 3 | 1 | 1 | 0 | 1 | 0 |
| Vespa mocsaryana | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 2 | 2 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| Vespa multimaculata | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| Vespa orientalis | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 0 | 1 | 0/1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| Vespa philippinensis | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| Vespa simillima | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| Vespa soror | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 0 | 3 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 1 | 1 | 0 | 1 | 0 |
| Vespa tropica | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| Vespa velutina | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| Vespa vivax | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
Morphological codes
(F / M): character restricted to Females / Males
1. Prestigma length: (0) prestigma shorter than pterostigma, (1) prestigma longer than pterostigma, (2) prestigma length 3X pterostigma length.
2. Base of second submarginal cell: (0) M obliquely oriented with respect to m-cu1, (1) M vertically oriented (angle at M noticeable).
3. Placement of forewing m-cu2: (0) close to r-m2, (1) far from r-m2.
4. Hamuli placement: (0) beginning basad of fork of R1&Rs, (1) beginning at fork R1&Rs.
5. Hindwing jugal lobe: (0) present, (1) absent.
6. Hindwing axillary incision: (0) present, (1) absent.
7. (M) Tyloides: (0) two on apical flagellomeres, (1) one on apical flagellomeres, (2) absent.
8. Vertex length: (0) ocelloccipital distance short, < length of ocellar triangle, (1) ocelloccipital distance long, > length of ocellar triangle, (2) ocelloccipital distance long, gena produced behind eye.
9. Vertex indentation: (0) absent, (1) present.
10. Ocelli diameter: (0) less than distance between posterior ocellus and eye, (1) greater than this distance.
11. Interantennal space: (0) broad, rounded, (1) defined triangular area, (2) strongly elevated, with rounded edges, (3) defined triangular area strongly elevated, with sharp edges.
12. Clypeus dorsum: (0) straight, (1) bisinuate.
13. (F) Apex of clypeus: (0) pointed, (1) shallowly emarginated, anterior angles blunt, broad, (2) shallowly emarginated, anterior angles triangular.
14. (F) Mesal clypeal tooth: (0) absent, (1) present.
15. (F) Clypeal punctures: (0) sparse, superficial mesally, (1) dense mesally.
16. (M) Clypeal-eye contact: (0) touching, (1) gap.
17. (F) Malar space: (0) short, (1) long, > length of penultimate flagellomere.
18. Mandibular teeth: (0) pointed, (1) with elongate cutting edge, twice length of apical part.
19. Labial palpus third segment: (0) with strong seta, (1) without this seta, but with hairs.
20. Pronotal carina: (0) present, (1) dorsally reduced, (2) lateral remnants, (3) absent.
21. Pronotal carina dorsally: (0) largely transverse before scutum, (1) deeply U-shaped before scutum.
22. Pronotal carina laterally: (0) little interrupted by the pronotal fovea, (1) widely interrupted by the pronotal fovea.
23. Pretegular carina: (0) complete, (1) ventrally effaced, (2) absent.
24. (F) Pronotal striae: (0) absent, (1) few ventral striae, (2) dense ventral and dorsal striae.
25. Scutal lamella: (0) present, (1) absent.
26. Scutal punctures: (0) dense micropunctures, (1) superficial, (2) dense and deep micropunctures.
27. Epicnemial carina: (0) present, (1) absent.
28. Dorsal groove: (0) present, (1) absent.
29. Scutellum profile in lateral view: (0) bulging, (1) largely flat.
30. Metanotal orientation: (0) partly vertical, (1) largely vertical (dorsal surface reduced).
31. Metanotal lobe: (0) absent, (1) posteromedial lobe present.
32. Metapleural sculpture: (0) striae, (1) superficial punctures ventrally, (2) well-defined punctures ventrally.
33. Hind coxa carina: (0) absent, (1) present.
34. Metasomal segment I: (0) rounded in lateral view, (1) sharply angled between anterior and dorsal faces.
35. Metasomal segment I length: (0) short, (1) long.
36. Metasomal tergum II lateral macropunctures: (0) superficial to sparse, (1) dense, well defined.
37. (M) Apical margin of metasomal sternum VI: (0) almost straight, (1) shallowly emarginated, (2) deeply emarginated.
38. (M) Apical margin of metqasomal sternum VII: (0) convex, (1) shallowly emarginated, (2) deeply emarginated.
39. (M) Median process of metasomal sternum VII: (0) absent, (1) present.
40. (M) Aedeagal apex: (0) little differentiated, (1) transverse, projecting laterally, (2) rounded with subapical processes, (3) spade-shaped, (4) elongate, (5) narrow, (6) subcircular, (7) triangular.
41. (M) Aedeagal apical lobes: (0) absent, (1) apex forming expanded lobes.
42. (M) Aedeagus width: (0) narrow throughout, (1) as wide or wider apically as medially, (2) narrower apically than medially.
43. (M) Aedeagal shaft: (0) non-bulbous, (1) bulbous.
44. (M) Digital apical processes: (0) absent, (1) present.
45. (M) Inner apical margin of paramere: (0) obtusely angled with aedeagus, (1) forming right angle to aedeagus.