(C) 2012 Katja C. Seltmann. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Hymenoptera exhibit an incredible diversity of phenotypes, the result of ~240 million years of evolution and the primary subject of more than 250 years of research. Here we describe the history, development, and utility of the Hymenoptera Anatomy Ontology (HAO) and its associated applications. These resources are designed to facilitate accessible and extensible research on hymenopteran phenotypes. Outreach with the hymenopterist community is of utmost importance to the HAO project, and this paper is a direct response to questions that arose from project workshops. In a concerted attempt to surmount barriers of understanding, especially regarding the format, utility, and development of the HAO, we discuss the roles of homology, “preferred terms”, and “structural equivalency”. We also outline the use of Universal Resource Identifiers (URIs) and posit that they are a key element necessary for increasing the objectivity and repeatability of science that references hymenopteran anatomy. Pragmatically, we detail a mechanism (the “URI table”) by which authors can use URIs to link their published text to the HAO, and we describe an associated tool (the “Analyzer”) to derive these tables. These tools, and others, are available through the HAO Portal website (http://portal.hymao.org). We conclude by discussing the future of the HAO with respect to digital publication, cross-taxon ontology alignment, the advent of semantic phenotypes, and community-based curation.
URI, morphology, biodiversity informatics
Hymenopterists share a common interest, in part, because their research almost invariably requires the study of or reference to hymenopteran anatomy. Which morphological characters are diagnostic or phylogenetically informative? How is behavior manifested morphologically? In what part of the body are certain genes expressed or certain odors detected? Our collective knowledge of this complex data source, however, remained decentralized, resulting in a corpus of observations that remained relatively disconnected and dispersed in the literature.
There have been numerous attempts to clarify relevant anatomy, spanning from comprehensive anatomical treatments of character systems across Hymenoptera (e.g.,
In 2006, at the 6th International Congress of Hymenopterists meeting in South Africa, a proposal was made to develop a new mechanism for clarifying hymenopteran anatomy: the Hymenoptera Anatomy Ontology (HAO;
The HAO has since grown into the largest, best illustrated, and most documented multi-species arthropod anatomy ontology and now includes 2055 anatomical concepts and 3622 terms for these concepts, which are extensively annotated (2880 image-based, sub annotated with 2912 figure annotations; 5686 text-based), with 269 references linking terms to concepts (see Table 1 for definitions of concept/term). These data are derived from existing publications (the HAO database contains >1000) and ongoing research by HAO curators and other contributors.
Glossary of ontology-related terminology.
| Word | Definition |
|---|---|
| Class | A synonym of concept, in the context of the HAO. |
| Concept | The idea (primarily the definition and related annotations) that circumscribes our understanding of an anatomical feature, as denoted by its structural properties. For example, “The tagma that is located anterior to the thorax” defines the concept commonly referenced by the term “head”. |
| Concept Drift | The application of a term to an increasingly diverse set of structures over time. |
| Genus-differentia | A type of definition structured so as to first describe a more inclusive class of concepts (genus) and then the characteristics differentiating (differentia) it from other children of that concept. Definitions in this format typically follow the pattern “The x that is y.” Biologists will recognize this as the way Linnaeus structured scientific names. |
| Homonym | A term that is used for two or more concepts. |
| Instance | A real-life exemplar of the concept. For example, the actual physical head of a specific specimen, perhaps one of many collected by Lubo Masner, is an instance of the HAO class identified by the URI http://purl.obolibrary.org/obo/HAO_0000397. |
| Label | The HAO has in the past used label and term interchangeably; here we use term to reflect the broader use of the latter. |
| Ontology | A set of concepts and relationships (properties) pertaining to a particular domain of knowledge (e.g. hymenopteran anatomy). Ontology is a mechanism of classification. In the case of the HAO, a goal is to allow a user to classify anatomical structures within a logically consistent framework. |
| Obsolete Concept | Once an HAO identifier (e.g. HAO:0000397) is assigned to an anatomical concept, that concept will not be altered except for misspellings or obvious grammatical errors. If there is a fundamental problem with a concept definition (and therefore its URI) that concept is made obsolete but remains resolvable. |
| Relationship | A property (or attribute) shared between two instances (e.g., “mouthparts part_of head”). Within the HAO, relationships are recorded as linking two concepts, however when the ontology is applied to real-life examples, the relationships apply to instances of those concepts. For example, “the eye is not part of the head” (as recorded in the ontology), but rather “my eye is part of my head”. |
| Semantic Phenotype | Structured annotations that represent observations of the phenome (see |
| Sensu | A data construct that is a combination of a concept, term, and an associated reference. For example, |
| Structural Equivalence | Topographical sameness of structures. |
| Synonyms | Two or more terms used to reference the same concept. For example, both terms “phallobase” and “paramere” have been used to reference the concept http://purl.obolibrary.org/obo/HAO_0000713. |
| Term | A name (e.g., word, phrase, acronym) representing a concept to a human user. For example, “head”. (compare label) |
| URI | In the context of the HAO, a unique identifier for a concept. HAO URIs are resolvable, i.e., if used in a web browser additional information will be provided to the person or agent (other computer) that made the request. For example, http://purl.obolibrary.org/obo/HAO_0000713 is the URI for the concept “The anatomical cluster that is composed of the cupulae, gonostipites and volsellae.” |
The HAO is more than a simple glossary, as it is formally structured and follows rules of logic (
The raw, archived representation of an ontology, such as the HAO, is an explicitly formatted and defined text file. Typically these files are in Web Ontology Language (OWL; http://www.w3.org/TR/owl-features) or Open Biomedical Ontology (OBO; http://www.geneontology.org/GO.format.shtmll) format. The OWL family of languages is a general-purpose means of representing knowledge on the Web, and their development is supported by the World Wide Web Consortium (W3C; http://www.w3.org/). OBO specifications are much more narrowly focused and technically simpler, their purpose being to serve the specific needs of biologists who use ontologies. With some exceptions not yet pertinent to the HAO, tools exist to losslessly (i.e., without loss of data/meaning) translate between OWL- and OBO-formatted ontologies, and the HAO can be downloaded in either format (OWL: http://purl.obolibrary.org/obo/hao.owl or OBO: http://purl.obolibrary.org/obo/hao.obo).
Where to find the HAOHymenopterists will most likely access the HAO through the HAO Portal website (http://portal.hymao.org/; see below), but the ontology itself is accessible through several widely used biomedical databases. For example, the HAO is one of many ontologies archived by the Open Biomedical Ontology (OBO) Foundry (http://www.obofoundry.org). The OBO Foundry supports archiving and development of OWL and OBO formats as part of an effort to maintain and promote the use of biological ontologies across biological and medical domains. The OBO Foundry also facilitates ontology dissemination and use, and ontologies archived there are automatically made available through other portals such BioPortal and Ontobee (for the HAO versions see http://bioportal.bioontology.org/ontologies/1362 and http://www.ontobee.org/browser/index.php?o=HAO respectively). Our association with the greater biomedical ontology community, especially the National Center for Biomedical Ontology (http://www.bioontology.org/), ensures that the HAO will be archived for long-term sustainability and distributed for broad use in other domains.
The HAO in the broader communityProtocols for ontology construction are produced by a large community of ontologists (HAO curators included), some of whom work with anatomy ontologies in practical ways and others who are more theoretical and primarily concerned with the logic inherent in the structure of ontologies. It is important for the HAO, as it is for any anatomy ontology, to be integrated into larger ontology initiatives. As contributors to the larger anatomy ontology community, the collective HAO is naturally incorporated into a field of research that embraces many biological disciplines and taxa. We benefit from, and contribute to, the efforts of the entire ontology community, including their infrastructure and software advances. By working within larger initiatives we ensure that the HAO is positioned to take advantage of ongoing initiatives (e.g. integration with the genomics community).
The HAO “ecosystem”The principal product of the HAO Project is the ontology itself, but the process of development has also resulted in multiple applications, built upon the ontology, that facilitate Hymenoptera research. HAO developers are responsible for creating human-usable Web interfaces and applications that make the text-file ontology into tools for the biological community, and to encourage an ontology-based approach to morphology. The primary application is the Hymenoptera Anatomy Ontology Portal (formerly the “Hymenoptera Glossary”). The HAO Portal displays data contained within the Hymenoptera Anatomy Ontology, as well as other information useful for Hymenoptera research including annotated images, alternative definitions, comments from users, ontology visualization tools, an extensive bibliography, and the “analyzer” tool (described below).
Unique identifiers for anatomical conceptsMost hymenopterists are familiar with the process of submitting sequences to GenBank and receiving unique identifiers (i.e., GenBank accession numbers) that serve as published references to their data. The HAO serves a similar function for anatomy by providing unique identifiers for anatomical concepts. The identifiers used by the HAO are Uniform Resource Identifiers (URIs) that consist of a Persistent Uniform Resource Locator (PURL: http://purl.obolibrary.org/obo/) plus an HAO identifier in OBO format (e.g. HAO:0000397). The latter is a combination of the namespace used in the OBO Foundry (“HAO”) and a unique seven-digit number (e.g. “0000397”). The “:” in the identifier is replaced with an “_” in the URL form. Any given URI points to only a single concept within the HAO (e.g. http://purl.obolibrary.org/obo/HAO_0000397 = “The tagma that is located anterior to the thorax” = head).
The virtue of URIsThe concept of the URI is important because it allows the HAO to be utilized in multiple contexts. Within Web browsers (e.g. Firefox, Google Chrome, Internet Explorer) many URIs (including the HAO’s) may be used in the same manner as URLs (Uniform Resource Locators) for webpages. URIs, in combination with Web-server configurations, allow for different responses, based on who is making the request. For example, a request from a Web browser—a person clicking on a link in a journal article or website—would return content that a human can interpret (e.g., a webpage), whereas requests from computational sources would receive responses that the application can understand (leaving out the human-readable components). This core functionality is at the heart of the long-term use and application of HAO URIs within publications, websites, applications and analyses. The challenge then is to facilitate the production, use, and publication of URIs (see below).
Referencing the HAO in publications The URI TableMaterials and methods sections within papers that reference hymenopteran anatomy frequently point to concepts published elsewhere (e.g., “Morphological terminology largely follows
An example URI table. Starred (*) columns are required. See discussion for details.
| Term* | URI* | Definition | Definition Source | HAO Preferred term | Comment |
|---|---|---|---|---|---|
| head | http://purl.obolibrary.org/obo/HAO_0000397 | The tagma that is located anterior to the thorax. | Snodgrass, R. E. 1935. | ||
| paramere | http://purl.obolibrary.org/obo/HAO_0000395 | The sclerite that is connected distally to the gonostipes. | Crampton, G. C. 1919. | harpe | |
| propodeum | http://purl.obolibrary.org/obo/HAO_0001249 | The area that is located posterior to the metapleural carina. | Mikó, I., L. Vilhelmsen, N. F. Johnson, L. Masner, and Z. Pénzes. 2007 | The propodeum here is also part_of the metapectal-propodeal complex- this further bounds the somewhat generalized defintion. |
1. Term: The literal text (string of letters) used in the paper for the anatomical concept. It is important to reference anatomical terms used within the table exactly throughout a paper, to eliminate ambiguity. Simplification of terms within a document, without including these simplifications in the table, reduces the effectiveness of a table intended to clarify terminology. For example, if “abdominal tergum 3” is in the table, the structure should be fully referenced throughout the document as “abdominal tergum 3” and not simplified to “the tergum” anywhere.
2. URI: The unique, resolvable, identifier for the anatomical concept. HAO curators create these identifiers, which can be found by accessing the HAO Portal.
3. Definition: A verbatim replication of the definition in the HAO, which are written as genus-differentia (see
4. Definition source: A citation from which the concept/definition was derived; within the HAO
5. HAO preferred term: The present preferred term for the concept denoted by the URI in the HAO (see below for explanation). This term is provided only when it does not match the term the author uses in his or her publication. This option provides authors the freedom to reference anatomy in any manner he or she sees fit.
6. Comment: Comments may pertain to any or all of the columns for a given concept. For example they can be used to clarify subtleties or provide taxon-specific discussion.
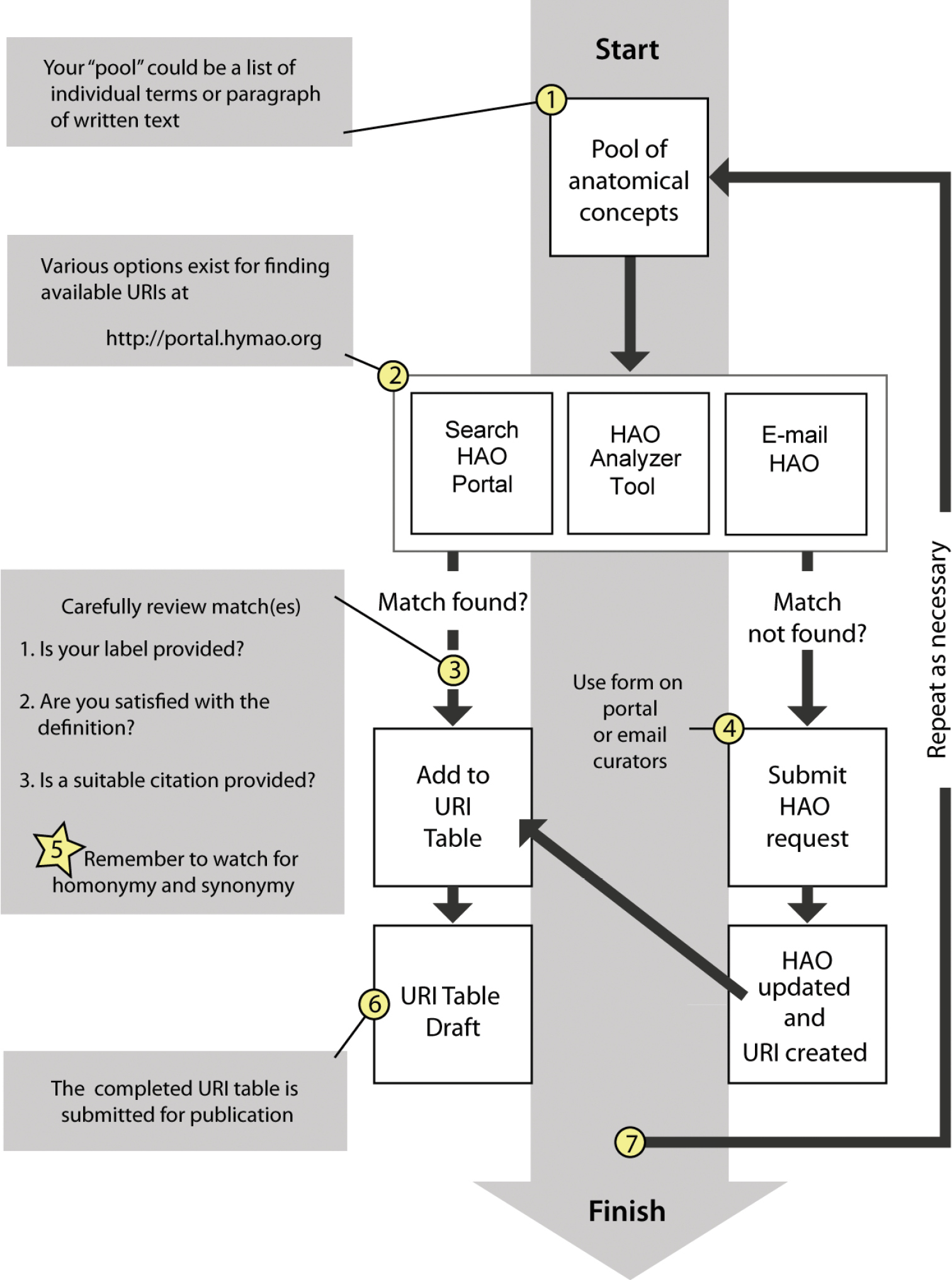
Creating a URI tableURI tables are created (Fig. 1) prior to submission of a manuscript, usually in close conjunction with an HAO curator, though they can also be built independently. The goal of building a URI table is to find or help create a URI that represents the anatomical structures referenced in a paper (though a URI table that only references a subset is still useful; see
Workflow for drafting a URI table.
The first step in building a URI table involves discovering available URIs (Fig 1:2), using tools available at the HAO Portal site: the simple Portal search tool, the “analyzer tool” (described in detail below), or by requesting help from the HAO e-mail list. If URIs are found they can be added to the table (Fig 1:3) after the user is satisfied with the HAO concept, citation, and labels provided. If a match is not found, or the author is not satisfied with a present concept, an HAO curator will work with the author to create a new URI (Fig 1:4). HAO curators can be contacted through the feedback form online (links from http://portal.hymao.org), through email directly to an HAO curator, or through contacting the broader HAO community on the listserv (http://purl.oclc.org/NET/hymontology/listserv). Authors are cautioned when reviewing concepts to watch for homonymy and synonymy, and not to assume that if a term match is returned that the definition will also match the authors notion of its meaning in the manuscript (Fig 1:5). In other words, authors should carefully review term definitions before adding them to the URI table. Tables can be included in Materials and Methods sections or as appendices, preferably as part of the manuscript rather than supplementary documents (Fig 1:6). While URI tables can be developed independently it is recommended that you pass along your table to the HAO curators to review prior to submission for publication. The entire process is iterative (Fig 1:7) and may be repeated multiple times as a manuscript develops.
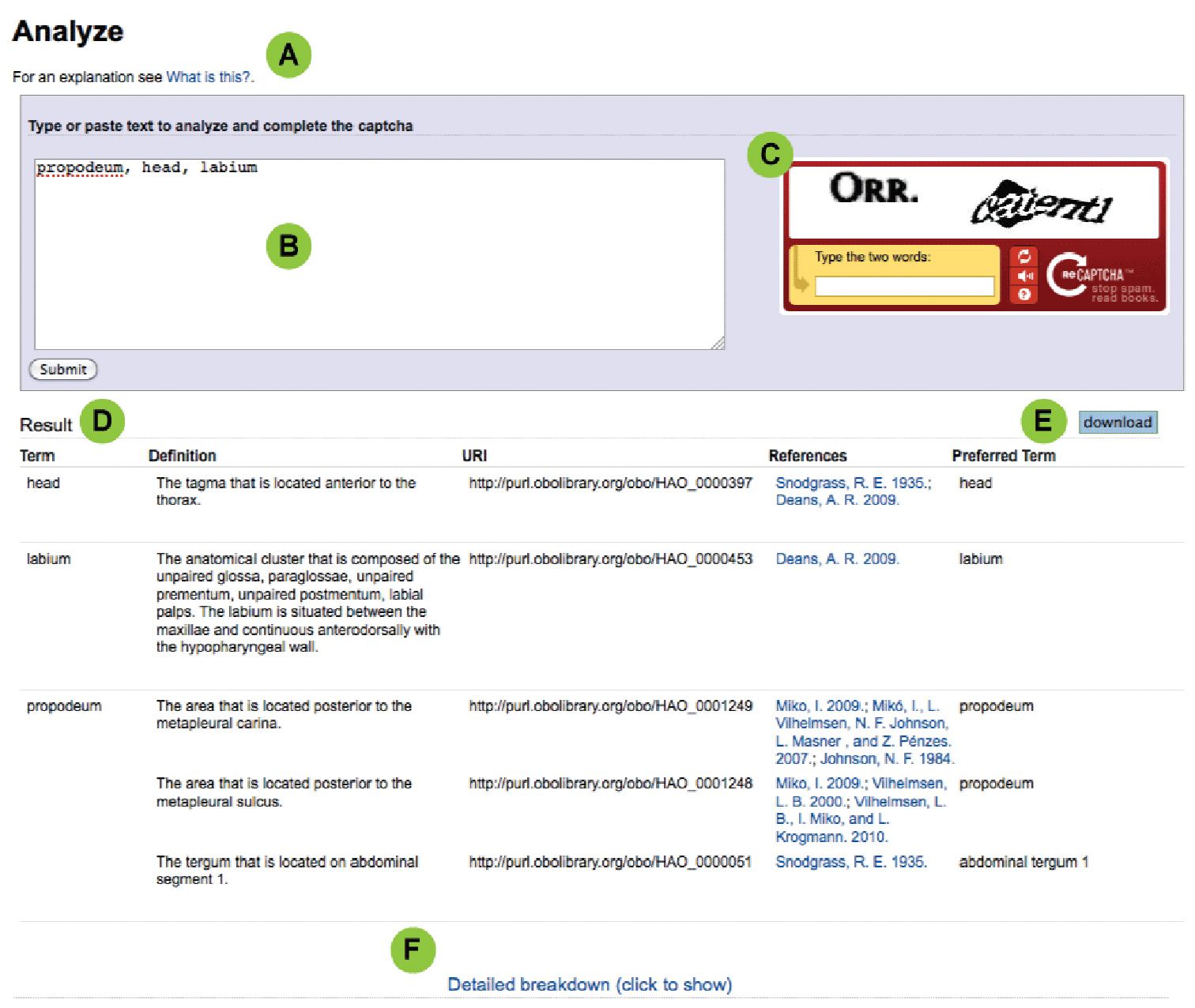
Using the “analyzer” toolThe goal of the analyzer tool (Fig 1:2 ) is to facilitate the adoption of URIs in publications and digital applications. The mechanism created to accomplish this is a simple interface (Fig. 2) for submitting an author’s anatomy-related text. The provided text is broken down into individual and groups of words, and these are compared against the terms in the HAO for matches by an internal algorithm. Terms are matched letter-for-letter, i.e., “forewing” will not match “fore wing”. No “fuzzy” matching attempts are made, i.e., the software does not try to predict what anatomical concepts the author is referring to. Matching terms are summarized by the anatomical concept to which they are attached (Fig. 2D). URIs for each matching concept are also provided. To use the analyzer follow these steps and see the online help (Fig. 2A, “What is this?”):
Using and interpreting results from the analyzer. See Using the “analyzer” tool for explanation. A Help link B Input field C CAPTCHA test D Result table E Download link; and F Detailed breakdown link.
1. Provide your text: Copy and paste your text, or type directly into the text box (Fig. 2B). Try to limit the text in the box to that which is pertinent to the underlying ontology being analyzed (e.g., do not include specimen examined sections from taxonomic descriptions, but rather the description alone). You can paste sentences, entire paragraphs from a manuscript description or diagnosis, comma-delimited lists, or any combination thereof. Typically, this would include a description and diagnosis from the manuscript.
2. Prove you are human: To discourage spammers and other troublemakers the analyzer form includes a “CAPTCHA” (http://www.captcha.net/) (Fig. 2C). Type the two words shown into the text box. If you need help click the small “?” on the form. Each time you click the submit button you will have to re-complete the CAPTCHA, even after you have successfully filled one out the first time.
3. Submit: Click Submit and wait (you will see a spinner turning). You might have to wait for a few seconds depending on the amount of text submitted.
4. Examine results: Results are displayed in a table (Fig. 2D) below the form. Be aware that if a term is not in the HAO it will not appear in the results list. Use the “Match Map” from the Detailed Breakdown Report (see Fig. 3) to check if all of your terms are included. References (Fig. 2, “References”) represent citations wherein the term was used in conjunction with the definition/concept as interpreted by an HAO curator. Carefully reviewing each conceptual match is critical; do not assume that the result is using the term you have provided in the manner you mean. Feedback based on missing terms, problems with definitions, etc. can be provided as outlined in Fig. 1 (and see Curators and Curation below). An additional breakdown of the results (Fig. 1F; Fig. 2) provides more details.
The detailed breakdown report from an analyzer report. See Using the “analyzer” tool for explanation.
5. Download results: Download the result by clicking the “Download” button (Fig. 1E). The downloaded file is a tab-delimited file that can be opened directly in a spreadsheet program (e.g., Microsoft Excel) or a text editor.
6. Edit the results: If multiple concepts (definitions) are available for a given term choose the one that matches your concept. If you need help determining which concept to select use a feedback mechanism (Fig. 1). Delete the other rows. Repeat for all the terms in the table.
7. See more: For additional details click on “Detailed breakdown” (Fig. 1F; Fig. 3). This report can be used to visually inspect for misformatted terms, alternate spellings or usages of a term, or potentially new additions to the HAO. The report includes:
a. Analyzed Text: Use this to confirm that your text was submitted correctly. The analyzer may truncate your text if it is too long, or if other problems are encountered.
b. Match Map: The words that were matched and returned are highlighted in the context of the text here. Green highlights indicate a 1:1 mapping (i.e., there is only one concept present for the given term). Pink highlights indicate that there are multiple concepts for a given term.
c. Matched Terms: A simple comma separated list of the terms that were found.
d. Matched Classes: A list of the concepts that were matched.
e. Homonymous Terms: Matched terms that are homonyms.
f. Synonymous Terms: Matched terms that are synonyms.
Recurrent IssuesThe HAO project is exemplary in its close connection to domain experts (hymenopterists), and its success depends on broad community support. During its short life, the potential utility of the HAO has already begun to emerge (e.g.,
Understanding the criteria for defining HAO concepts is important for understanding the decision-making process for constructing the HAO and how it can be useful as a tool for referencing and aligning anatomical concepts. Fundamentally, the HAO project rests on recognizing different instances of a topographically-defined concept as “the same” (e.g., the fore wing of taxon A is the same structure as the fore wing of taxon B) (topographical homology of
The HAO provides structure-based anatomical concepts, from which homology hypotheses can be developed and subsequently tested. This strategy derives from the premise that statements of homology, in the absence of phylogenetic tests, should be avoided. This is not to say that the construction of the HAO is not influenced by (and can influence) phylogeny-based homology hypotheses. New concepts can be derived as necessary, but these concepts will be rooted in what can be observed, not in what is hypothesized. One of the HAO’s greatest strengths is that it forces the careful introspection necessary for generating strong primary homology hypotheses by forcing users to reconcile a localized understanding (e.g. an author’s unwritten understanding of the anatomical nuances) with a broader conceptual framework. Many HAO concepts are sufficient for the basis of evolutionary homology hypotheses; however this is not always the case.
One example of how a structurally equivalent concept might not be homologous pertains to processes (http://purl.obolibrary.org/obo/HAO_0000822). Many distantly related hymenopterans have similarly located (i.e. structurally equivalent) pronotal spines. These spines would correspond to the same concept in the HAO, i.e., they are analogous but are not necessarily homologous.
Classifying anatomical structures using structural equivalencyStructural equivalency allows clustering of anatomical structures described in the literature, based on clear, repeatable criteria—i.e. the criteria are observable, not hypothesized. Because of the structural equivalency criterion, the match between ontology concept and author concept of a term does not need to be exact but rather only structurally equivalent. HAO definitions, then, do not include all of the nuances of a concept; only the author can articulate those. When an author’s concept of a term (based on observations pertaining to its structure) differs from that of the definition in the HAO (including its properties) a new concept should be created.
If an author intends to state that a particular anatomical concept was observed in some instance of a hymenopteran then that author must ensure that they observed all of the components of the anatomical concept provided in the HAO for that concept. For example, in many HAO concepts points of muscle attachment are used (an important line of evidence for topographical definitions). The author must have observed these muscles and their points of attachment to state that the class in question was present in their hymenopteran instance. Careful attention must also be made to the relationships a particular anatomical concept has to other concepts; these relationships must also be observed by the author.
There is, however, one circumstance in which anatomical concepts in the HAO might be “loosely referenced” (e.g., referencing an anatomical concept that includes, in part, reference to muscle arrangements when said muscle arrangements have not been observed). This is during the formulation or discussion of hypotheses, often pertaining to character evolution. For example, a paleontologist studying two fossils may wish to discuss them in the context of anatomical concepts in the HAO: “I hypothesize that fossil one bears anatomical concept HAO 1234 except for feature A (which I cannot observe based on preservation), and fossil two bears anatomical concept HAO 1234 except for feature B (which I believe is absent, but I would like to see more specimens)”. In this hypothetical scenario minting two new HAO URIs (i.e., HAO 1234 minus feature A, HAO 1234 minus feature B), gains us little (and may be positively misleading based on the uncertainty of the observer), in the absence of additional observations. The referencing to HAO 1234, however, provides an anchor to a discussion of the evolution of an anatomical concept, something potentially of use to future researchers.
Separation of terms and conceptsTerms and concepts are treated separately, and only concepts are given unique identifiers. Terms in the HAO are considered to be simply strings of letters that are used to reference concepts, associated together because they are structurally equivalent anatomical structures. Concepts encapsulate meaningful anatomical observations. Different observers looking at the same anatomical structure can have the same conceptualization of it, where it is located on the body, and the circumscription of area around it. Yet even with this similarity, they may still use different terms for the observed anatomical structure (synonymy). Different observers may use the same term yet have different understandings of when and how that term is applicable (the “sensu” construct of
Because there are no international rules of anatomical nomenclature, as there are for zoological nomenclature, anyone can use any terms they want to refer to an anatomical concept of interest. The HAO stands as a robust mechanism (through URIs) to clarify diverse concepts. Given the dual legacies of published literature and training histories, we envision a community where personalized lexicons continue to be refined and accommodated. Many hymenopterists, however, including a group at the 7th International Congress of Hymenopterists (Kőszeg, Hungary; June 2010), have called for the HAO Project to serve as an instrument for deciding upon preferred terms for concepts. Preferred terms facilitate accurate communication by simplifying concordance of terms with concepts, and they are required by potential HAO users in biomedicine.
Terms in a manuscript are strings of letters that may or not have meaning to a reader depending on his or her knowledge. A reader experienced enough with the terminology typically used to describe a certain taxon will not necessarily need to check the attached URI table to understand the meaning of a term. For a non-expert reader, unfamiliar with terms in a manuscript, preferred terms may be meaningless (or even misleading). However, preferred terms are very important for experts, who use “short” terms to encompass a hidden mountain of unexpressed background information. Preferred terms also are of use to taxonomists in the early stages of developing a vocabulary for their taxa, as the set is much smaller than the total number of possibilities. If an aspiring taxonomist can navigate directly to this set they potentially become more efficient in its use and less confused by the historical baggage carried by a larger set of overlapping terms.
The HAO now has an accessible listserv (http://purl.oclc.org/net/hymontology/listserv) for preferred term voting. Preferred term choices are emailed to the list once or twice a week and the votes are collected with the term obtaining the most votes moved into the “preferred” position on the HAO Portal webpages. All other terms are then considered synonyms. The “term of the week” is selected based on the frequency of term usage found in past issues of the Journal of Hymenoptera Research. As of this writing, listserv members have voted on about 60 preferred terms, all of which may be revisited by calling for a new discussion on the listserv.
Why not simply include definitions in one’s paper?Ontologies have structure, and concept definitions therefore inherit the definitions of all the concepts contained within the definition and the concepts it is related to. Authors themselves cannot easily explicitly define all the terms used in a publication, as their definitions will inevitably include terms that must also be defined. To precisely define one concept requires an entire ontology and one that is internally consistent (i.e. only uses terms that are also defined in the ontology). In addition, the HAO project provides a wealth of additional information. By linking to concepts in the HAO authors not only explicitly define terms in their publications, but the reader also gets other information about the concept, such as which major publications used the concept, its synonyms, homonyms, annotated images, and additional comments from the HAO curators.
PersistenceOur ideas of concepts may change over time, but the original concepts will never disappear in HAO. Once an HAO identifier (e.g. HAO:0000397) is assigned to an anatomical concept, that concept will not be altered except for misspellings or obvious grammatical errors. If there is a fundamental problem with a concept definition (and therefore its URI) that concept is made obsolete. Links to the obsolete concept persist, however, so that published URIs will always resolve, regardless of their standing in the current HAO. Obsolete classes in the HAO require statements as to why they were made obsolete and links to the newly derived concepts where pertinent. The obsoletion mechanism, in conjunction with a pluralistic approach to providing concepts (i.e., it is not a problem to create subtly different concepts), are strategies to minimize concept drift.
Web-browsersSome functionality of the HAO Portal currently does not work for some Web browsers. The HAO Project strongly recommends using Firefox (http://firefox.org) to view the HAO Portal pages in order to assure the proper rendering of annotated images. The HAO Portal utilizes cutting-edge functionality that some browsers (e.g., Internet Explorer, IE) presently do not support. However, if a person accesses a webpage in IE a warning message will appear directing the user to open the page in Firefox. Because free, standards-compliant browsers are available we do not see this requirement as a critical limitation. We anticipate that advances in browser capabilities related to the advent of the HTML5 standard will ensure that the HAO Portal will shortly truly become cross-browser compatible.
The Future of the HAOThe HAO is a resource based on a foundation of explicitly defined anatomical concepts and a straightforward mechanism for dereferencing these concepts (URIs). This resource is intended for any and all users who reference Hymenoptera anatomy. The HAO, like other biological ontology efforts, is rapidly evolving, both in its underlying data and its application. Its adoption depends first and foremost on the support of the hymenopterist community. The HAO is poised to succeed thanks, in part, to technological advances in publication, ontology alignment, and the burgeoning potential of semantic phenotypes (
Publications that precisely illustrate morphology routinely become the most cited and useful papers for future hymenopterists. The Journal of Hymenoptera Research (JHR) serves many experts and is well positioned to benefit from the rapidly growing publishing tool kit. The open-access model adopted by the JHR embraces the Web, pushing publications out of the traditional print-only format. Additionally, the JHR is published by Pensoft Press, which uses the TaxPub XML schema (
As ontologies continue to be adopted beyond the model-organism community they will be increasingly cross-referenced. Precisely defined concepts for hymenopteran anatomy, for example, can be connected to anatomical concepts in other domains that also have an ontology (e.g. Diptera). These alignments have the potential to facilitate novel evolutionary developmental biology hypotheses and hypotheses of homology. For example, while aligning the muscles in the FlyBase anatomy ontology (Drosophila; http://flybase.org/) with those concepts in the HAO, HAO developers noticed that the muscle patterns of the hymenopteran meso- and metathorax share similarities with the metathorax of different Drosophila Ultrabithorax (Ubx) mutants (see
The process of alignment and integration across ontologies is itself an intellectually challenging endeavor. Work facilitated by the Phenotype RCN (http://www.phenotypercn.org), which includes several HAO developers, is supporting the development of new arthropod-specific ontologies (e.g. one each for Coleoptera, Neuroptera, and Arthropoda). Work of this nature and scope is novel and will undoubtedly lead to big discoveries and new interpretations.
Semantic phenotypesNo formal organization currently governs the HAO, but the International Society of Hymenopterists has extended their support in making sure its future is secure. An ontology working group was formed (see ISH newsletter, March 2006), and the Society has offered to host HAO resources through their website should that service be desired by the community. The de facto curators (IM, MJY, ARD, KCS, MAB) strongly support community input and governance and will work with stakeholders to develop mechanisms for recruiting other curators and for facilitating decisions about ontology development (which must follow the recommendations of the broader ontology community, mainly the National Center for Biomedical Ontology and the OBO Foundry principles, to ensure robustness and broad use).
ConclusionIn addition to increasing the repeatability of our research, references to well-defined and illustrated anatomical concepts will open up their interpretation and use to a much broader array of users than just highly specialized scientists. Biodiversity, host-parasite biology, collections digitization, genomics, ecology, evolutionary developmental biology (evo-devo), invasive species evaluation, agro-ecosystem management and biological control (to name just a few) as well as those aspects of society impacted by these all rely on the correct interpretation of anatomical structures (e.g.,
This work was made possible by the U.S. National Science Foundation, grants DBI-0850223 and DEB-0842289, by a grant from the Encyclopedia of Life’s Biodiversity Synthesis Center (BioSynC), and by the Phenotype Research Coordination Network (NSF DEB-0956049). USDA is an equal opportunity provider and employer.